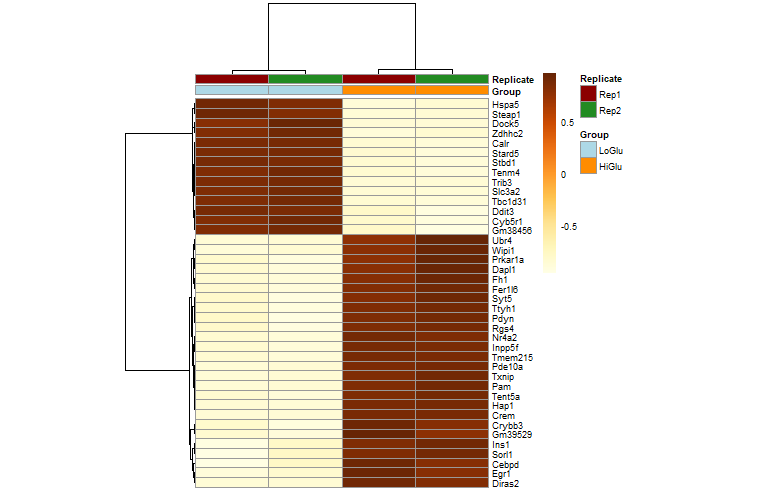
library(pheatmap)
library(RColorBrewer)
# 转换到rlog
dds_rlog <- rlog(dds, blind=FALSE)
# 获得前40差异基因
mat <- assay(dds_rlog[row.names(diff_gene)])[1:40, ]
# 选择用来作图的列
annotation_col <- data.frame(
Group=factor(colData(dds_rlog)$Group),
Replicate=factor(colData(dds_rlog)$Replicate),
row.names=colData(dds_rlog)$sampleid
)
# 修改颜色
ann_colors <- list(
Group=c(LoGlu="lightblue", HiGlu="darkorange"),
Replicate=c(Rep1="darkred", Rep2="forestgreen")
)
# 画图
pheatmap(mat=mat,
color=colorRampPalette(brewer.pal(9, "YlOrBr"))(255),
scale="row", # Scale genes to Z-score (how many standard deviations)
annotation_col=annotation_col, # Add multiple annotations to the samples
annotation_colors=ann_colors,# Change the default colors of the annotations
fontsize=6.5, # Make fonts smaller
cellwidth=55, # Make the cells wider
show_colnames=F)